 |
de | fr | en print view![]()
3R-INFO-BULLETIN 39
February 2009
Authors
 Dr. Paolo Cinelli is the head of the Molecular Genetics Laboratory at the Institute for Laboratory Animal Science of the University of Zurich. He has successfully assessed pain by telemetry in various biopsy sampling methods for genotyping. Igor Asner is currently working for his PhD on the assessment of pain by monitoring gene expression.
Dr. Paolo Cinelli is the head of the Molecular Genetics Laboratory at the Institute for Laboratory Animal Science of the University of Zurich. He has successfully assessed pain by telemetry in various biopsy sampling methods for genotyping. Igor Asner is currently working for his PhD on the assessment of pain by monitoring gene expression.
Address:
Paolo Cinelli
paolo.cinelli@ltk.uzh.ch
Igor Asner
igor.asner@access.uzh.ch
Institute of Laboratory Animal Science, University of Zurich
CH-8057 Zurich, Switzerland
Editor
Peter Maier, Scientific Adviser of the 3R Research Foundation
Detection of Pain in Laboratory Animals via Gene Expression?
Pain[*] assessment in animal experiments is a prerequisite for the appropriate use of analgesics. In order to develop a simple biochemical pain detection test, the expression levels of possible pain associated genes was investigated in a surgical pain model. In this project (nos. 96-05), supported by the 3R Research Foundation, initial exploratory investigations were carried out. So far they were without straightforward results, but data might serve as a base for the development of a pain detection test in the future.
Proper pain assessment
Mice are very commonly used as animal disease model. They live in constant fear of their predators and therefore show only discrete behavioural changes which reveal that they are suffering. An additional uncertainty derives from the observer’s subjective judgments during pain assessment. Telemetric measurements revealed that pain (e.g. after telemetry implantation) increases the heart rate (1) at least for 3 days after surgery and remain when analgesics (e.g Flunixin) are administered. Increase of heart rate can therefore be used for the determination of pain level, for example, when biopsies are taken for genotyping (2). However noninvasive and reliable pain assessment methods are required for routine use in animal experiments. In the present study telemetry apparatus implantation itself was used as a surgical pain model. Fluxinin was chosen as a postoperative analgesic, also given to control animals without surgery.
Pain perception controlled by gene products?
Individual differences in the perception of pain have often been described, and the question of a genetic basis to pain is a growing field of research. Many transgenic mice strains have been generated, which either exhibit allodynia or insensivity to pain (3). Moreover, changes in gene expression levels have been shown in rat pups, which had been repeatedly subjected to a painful stimulus. This genetically determined different response might explain why these pups remain less sensitive to pain into adulthood (4, 5). Other studies in rats have showed, that an inflammatory stimulus in adult rats leads to a long lasting hyperalgesia, and gene expression changes in the spinal cord (6). Accordingly it could be postulated that genes known to be involved in pain might show changes in their expression levels during or after a noxious stimulus.
More than 200 genes assessed
Microarray technology allows the simultaneous measurement of the expression levels of a large number of genes. We designed a microarray to analyze 221 genes for which studies showing that they are involved in pain mechanisms have been published (Table 1).
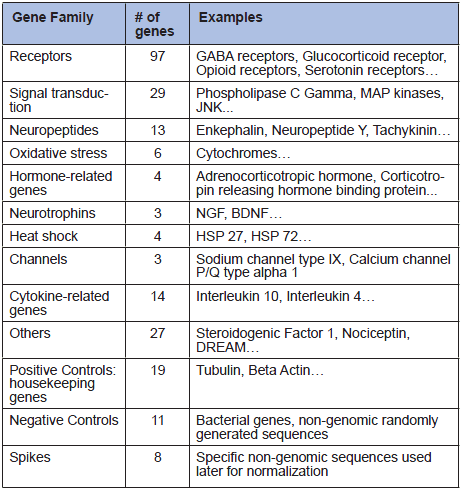
Table 1
Gene families analyzed in microarray analysis.
In the first set of experiments, gene expression changes in the whole brain and the spinal cord were measured in mice which had undergone a telemetry implantation. Control mice received the same anesthesia, but did not undergo surgery. No significant difference in gene expression was observed for the selected genes in the brain or the spinal cord. It was argued that a gene expression analysis in the whole brain or the whole spinal cord is not sensitive enough, because gene expression changes which happen only in defined parts of the central nervous system remain undetectable, as they get compensated by normal expression levels in the rest of the central nervous system.
Brain region specific expression?
Accordingly, we decided to analyze gene expressions levels in different regions of the brain (Figure 1). The same surgical design was applied as in the previous experiment. After sacrifice, the brainstem, the cortex, the hippocampus, the amygdala and the cerebellum were dissected from each brain, and the RNA from each part was analysed. In the 3 operated animals and the 3 control animals, no significant differences could be observed. However, we detected differentially expressed genes between the various parts of the brain. Our experiments showed 37 genes differentially expressed between the cerebellum and the brainstem, 43 genes were differentially expressed between the cerebellum and the hippocampus, 26 genes between the brainstem and the hippocampus, and 26 between the amygdala and the brainstem (ANOVA tests, p +/-0.05 for each gene). This data was consistent with previous published microarray data comparing the same parts of the brain (7). Our microarray and our protocol can therefore be validated as a suitable tool to observe gene expression differences in the brain, but might not be sensitive enough to measure changes due to pain.
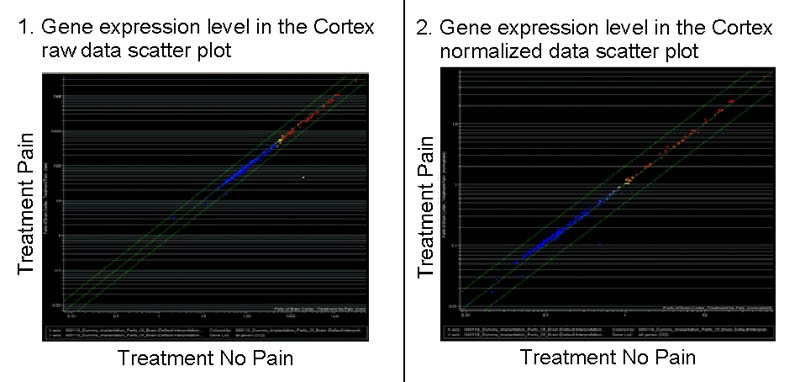
Fig. 1
Scatterplots of gene expression levels in the cortex.
The x-axis indicates the fluorescence intensity (gene expression) in the control groups and the y-axis in the operated animals. Left, a scatter plot of the raw data directly from the scanner and right, data normalized with the expression levels of positive control genes. In both cases, all the genes lie in the midline, indicating no relevant expression level changes between the operated animals and the control ones.
More specific methods required?
Real-Time PCR is a more specific method than microarray to investigate gene expression, as specific primers and annealing temperature are used for each gene. However, fewer genes can be analyzed in one run. We carried out some Real-Time PCR analyses on a set of 27 genes from our array, selected because of their relevancy in pain research (Table 2). The same surgical process as in the previous experiments was followed. After 2 days, we dissected the cortex, the hippocampus, the brainstem, the cerebellum, the spinal cord and 6 lumbar dorsal root ganglia from each animal. No significant expression level changes could be observed between the operated animals and the controls in the dissected 5 brain tissues (Figure 2).
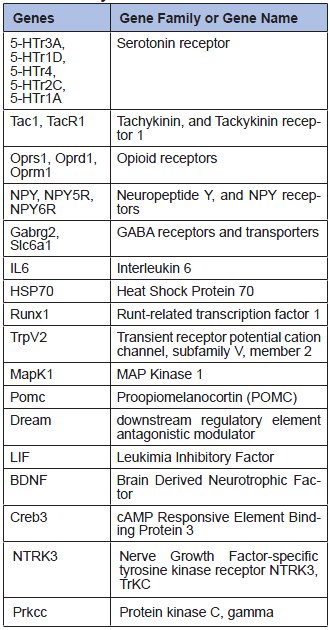
Table 2
27 genes analyzed by Real-Time PCR.
In order to see if the genes show expression levels changes in another type of pain, we conducted a parallel experiment in an inflammatory pain model (intraperitoneal injection of Complete Freund’s Adjuvant). The spinal cord RNA was analyzed by Real-Time PCR, and again no significant gene expression differences between the two groups could be observed. This suggests that the expression level of these 27 genes remains unchanged between animals that experienced surgical or inflammatory pain and control animals.
Unknown gene products involved?
Microarray or Real-Time analysis of gene expression could pave the way for the development of pain-detection tests based on gene expression. Sentinel animal could be assessed prior to experiments to detect the pain level of a procedure. So far our data have indicated that none of the commonly known “pain genes” show a differences in their expression levels in different brain areas. We are currently performing whole genome analyses of mice, which underwent the surgical protocol as mentioned above. By that we hope to identify new candidate genes, which show differences in their expression levels, and could be efficiently used to monitor pain in laboratory animals.
PDF version of this Bulletin No. 39
References:
- Arras, M., Rettich, A., Cinelli, P., Kasermann, H.P., and Burki, K. (2007). Assessment of post-laparotomy pain in laboratory mice by telemetric recording of heart rate and heart rate variability. BMC veterinary research 3, 16.
- Cinelli, P., Rettich, A., Seifert, B., Burki, K., and Arras, M. (2007). Comparative analysis and physiological impact of different tissue biopsy methodologies used for the genotyping of laboratory mice. Lab Anim 41, 174-184.
- Mogil, J.S., and McCarson, K.E. (2000). Identifying pain genes: bottom-up and top-down approaches. J Pain 1, 66-80.
- Anseloni, V.C., He, F., Novikova, S.I., Turnbach Robbins, M., Lidow, I.A., Ennis, M., and Lidow, M.S. (2005). Alterations in stress-associated behaviors and neurochemical markers in adult rats after neonatal short-lasting local inflammatory insult. Neuroscience 131, 635-645.
- Barr, G.A., Gao, P., Wang, S., Cheng, J., Qin, J., Sibille, E.L., and Pavlidis, P. (2005). Microarray analysis of gene expression following the formalin test in the infant rat. Pain 117, 6-18.
- Yukhananov, R., and Kissin, I. (2008). Persistent changes in spinal cord gene expression after recovery from inflammatory hyperalgesia: a preliminary study on pain memory. BMC neuroscience 9, 32.
- Hovatta, I., Tennant, R.S., Helton, R., Marr, R.A., Singer, O., Redwine, J.M., Ellison, J.A., Schadt, E.E., Verma, I.M., Lockhart, D.J., et al. (2005). Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature 438, 662-666.
| [*] | The mechanisms of Pain.Pain is commonly seen as an unpleasant experience, which signals a tissue injury, and is in most cases generated by a noxious stimulus. It is essential for the survival and the development of organisms, and it lies on the complex interaction of physiological and psychological mechanisms. Three different types of pain are commonly described: nociceptive pain, inflammatory pain, and neuropathic pain. Nociceptive Pain is caused by an exterior stimulus, such as noxious heat, noxious cold, and mechanical or acidic stress. Each of these stimuli activates a specific set of receptors at the peripheral terminals of the Aδ and C fibers. If the stimulus is intensive enough, the message will be transferred to the spinal cord where it will be processed. The first process will be a reflex message which will return to the periphery, and will, for example cause a withdrawal reflex reaction. The second process is the transmission of the message to the brain. The message is transmitted to the thalamus and the somatosensory cortex, which are involved in the painful sensation and to the brainstem, the hippocampus and the amygdala, which are involved in memory and emotions, and are therefore responsible for the learning and affective processes associated to pain. Inflammatory pain is due to the release of specific substances by the immune system cells, at a site of injury. These substances modify the excitability of nociceptors. Therefore a stimulus, which normally would not be noxious will become painful. Neuropathic pain happens after an injury in the nervous system. It is characterized by a series of inflammatory processes and structural changes which lead to a persistent form of pain. |
| Last modified 2009/03/28 |